Supra adjacency matrices¶
Multiplex (layer) networks can also be represented as supra-adjacency matrices as follows:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 | ### simple supra adjacency matrix manipulation ## tensor-based operations examples from py3plex.core import multinet from py3plex.core import random_generators ## initiate an instance of a random graph ER_multilayer = random_generators.random_multilayer_ER(500,8,0.05,directed=False) mtx = ER_multilayer.get_supra_adjacency_matrix() comNet = multinet.multi_layer_network(network_type="multiplex",coupling_weight=1).load_network('../datasets/simple_multiplex.edgelist',directed=False,input_type='multiplex_edges') comNet.basic_stats() comNet.load_layer_name_mapping('../datasets/simple_multiplex.txt') mat = comNet.get_supra_adjacency_matrix() print(mat.shape) kwargs = {"display":True} comNet.visualize_matrix(kwargs) ## how are nodes ordered? for edge in comNet.get_edges(data=True): print(edge) print (comNet.node_order_in_matrix) |
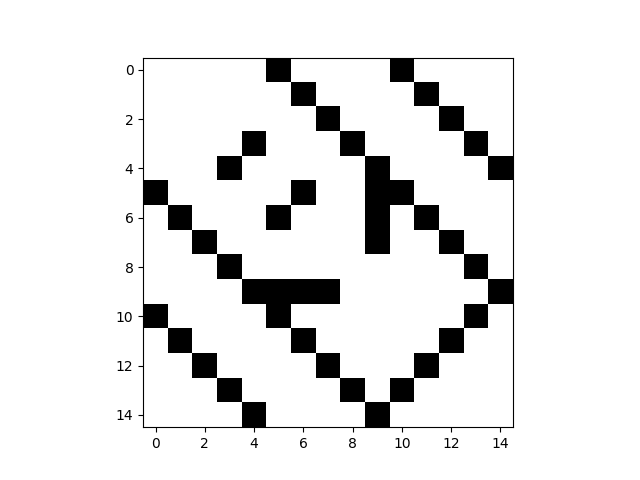
Some additional tensor-like indexing:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 | ## tensor-based operations examples from py3plex.core import multinet from py3plex.core import random_generators ## initiate an instance of a random graph ER_multilayer = random_generators.random_multilayer_ER(500,8,0.05,directed=False) ## some simple visualization visualization_params = {"display":True} ER_multilayer.visualize_matrix(visualization_params) some_nodes = [node for node in ER_multilayer.get_nodes()][0:5] some_edges = [node for node in ER_multilayer.get_edges()][0:5] ## random node is accessed as follows print(ER_multilayer[some_nodes[0]]) ## and random edge as print(ER_multilayer[some_edges[0][0]][some_edges[0][1]]) |